Protein blocks are structural prototypes defined by de Brevern et al. [1] (see also references [2] & [3]). Their main interest is to modelize a 3-dimension local structure into a 1-dimension sequence. In principle, any conformation of any amino acid could be represented by one of the sixteen available protein blocks (Figure 1).
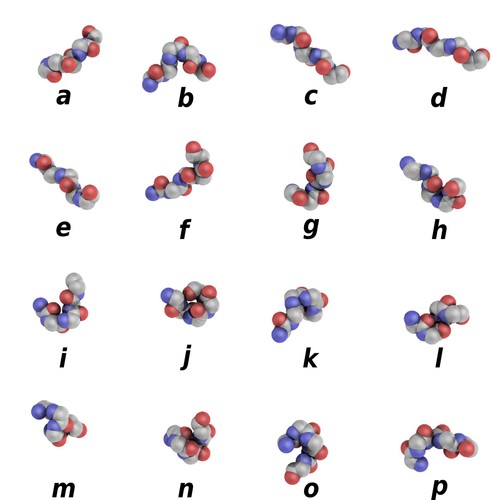
Figure 1. Schematic representation of the sixteen protein blocks, labeled from a to p. (Creative commons CC BY). Larger picture available here
In the following example, the 3D structure of the barstar protein in Figure 2 is expressed in term of protein blocks:
ZZdddfklpcbfklmmmmmmmmnopafklgoiaklmmmmmmmmpacddddddehkllmmmmnnomm
mmmmmmmmmmmmnopacddddZZ
The conformations of the 89 residues are translated into a sequence of 89 protein blocks. Note that "Z" is a default name given to amino acid for which a protein block cannot be assigned. Indeed, the assignation procedure for a given residue also requires the conformation of the two residues placed before and the two residues placed after the residue under consideration. Therefore, a protein block cannot be assigned to the two first (N-termini) and two last (C-termini) residues of a polypeptide chain.
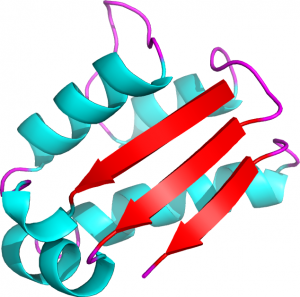
Figure 2. 3D representation of the barstar protein (PDB ID 1AY7, chain B) (Creative commons CC BY)
References:
[1] A. G. de Brevern, C. Etchebest and S. Hazout Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks Proteins 41: 271-287 (2000). HAL
[2] A. G. de Brevern New assessment of a structural alphabet In Silico Biology 5: 283-289 (2005). HAL NCBI
[3] A. P. Joseph et al. A short survey on protein blocks Biophysical Review 2: 137-145 (2010). HAL NCBI